Training a deep learning model for single-cell segmentation without manual annotation
Takeaways
-
The paper proposes a loss function to train neural networks (e.g. U-net) for single-cell image segmentation without labeled data (i.e. unsupervised learning).
-
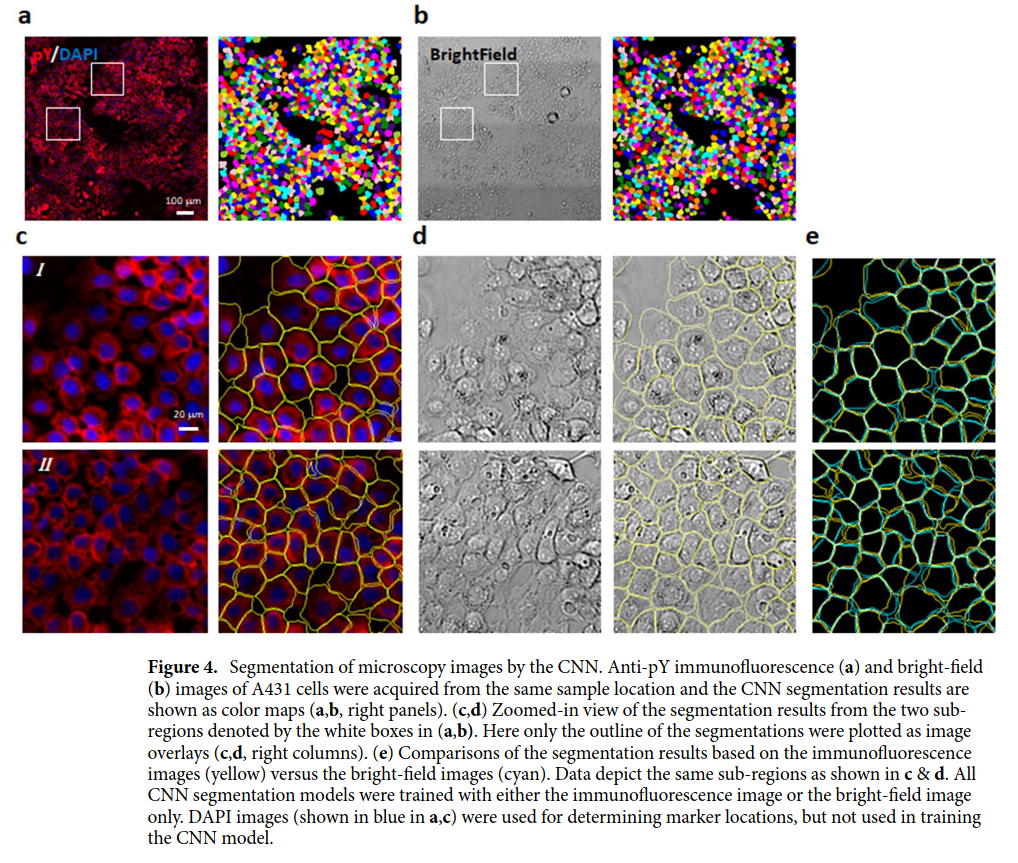
The input images (see figure above) are from 2D cell cultures (i.e. cell monolayers). The cells are generally packed together, but some empty spaces between them may occur.
-
The method requires knowing beforehand the position of the cell’s centers and where empty spaces (i.e. the background) lie. Therefore, although the technique is said to be unsupervised, it requires some labeled data. Nevertheless, the paper uses techniques to get these labels automatically. No manual labeling is required.
-
The cell’s nucleus location is used as an approximation of the cell’s center. The position of the nucleus is determined from images stained with DNA binding dye DAPI.
-
The background is segmented with a traditional algorithm (graph-cut).
-
During training, the full microscopy image is divided into small patches centered at each cell nucleus. The patch is just big enough to display the entirety of a single cell. The objective of the network is to segment only that single cell.
- The loss function can be expressed as the weighted sum of three costs:
- Overlapping segmentation of different cells. This makes sense. The cells are distributed in a monolayer, so indeed there is no overlapping.
- Cell segmentation invading the background region. Again, the background segmentation is known a priori (via graph-cut).
- Segmenting cells with a small area. This cost is especially required to avoid the trivial solution of having no cell segmented at all.
- The paper reports an mIOU of 82.1% on their own dataset.